Allosteric
Thank you for visiting nature. You are using a browser version with limited support for CSS, allosteric.
These examples are programmatically compiled from various online sources to illustrate current usage of the word 'allosteric. Send us feedback about these examples. Accessed 4 Mar. Subscribe to America's largest dictionary and get thousands more definitions and advanced search—ad free! See Definitions and Examples ». Log In. Examples of allosteric in a Sentence.
Allosteric
Thank you for visiting nature. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser or turn off compatibility mode in Internet Explorer. In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript. Allostery in proteins influences various biological processes such as regulation of gene transcription and activities of enzymes and cell signaling. Computational approaches for analysis of allosteric coupling provide inexpensive opportunities to predict mutations and to design small-molecule agents to control protein function and cellular activity. We develop a computationally efficient network-based method, Ohm, to identify and characterize allosteric communication networks within proteins. Unlike previously developed simulation-based approaches, Ohm relies solely on the structure of the protein of interest. We use Ohm to map allosteric networks in a dataset composed of 20 proteins experimentally identified to be allosterically regulated. Our webserver, Ohm. David Ding, Ada Y.
Role of conformational sampling in computing mutation-induced changes in protein structure and stability, allosteric.
Federal government websites often end in. The site is secure. Allosteric drugs are currently receiving increased attention in drug discovery because drugs that target allosteric sites can provide important advantages over the corresponding orthosteric drugs including specific subtype selectivity within receptor families. Consequently, targeting allosteric sites, instead of orthosteric sites, can reduce drug-related side effects and toxicity. On the down side, allosteric drug discovery can be more challenging than traditional orthosteric drug discovery due to difficulties associated with determining the locations of allosteric sites and designing drugs based on these sites and the need for the allosteric effects to propagate through the structure, reach the ligand binding site and elicit a conformational change.
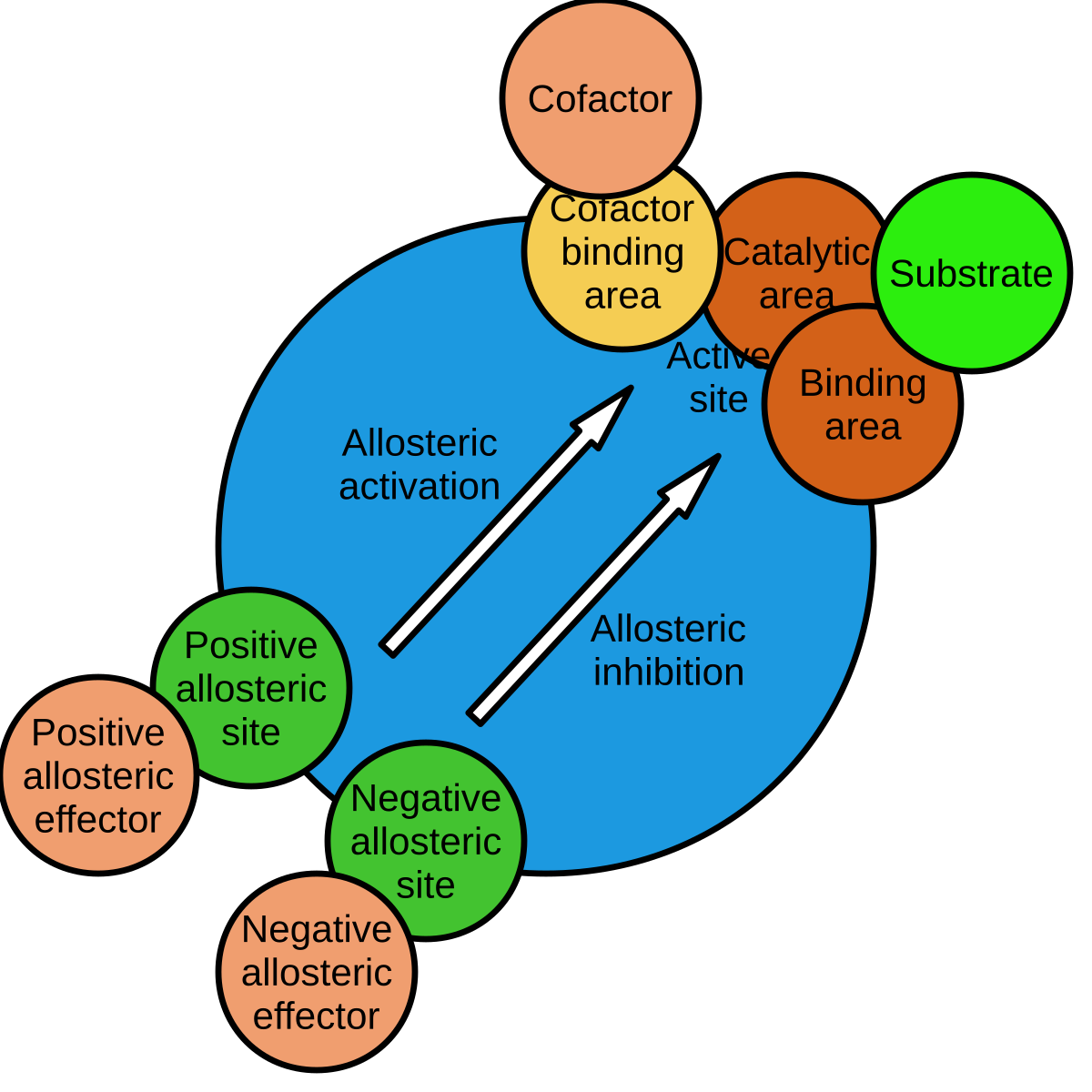
In pharmacology and biochemistry , allosteric modulators are a group of substances that bind to a receptor to change that receptor's response to stimuli. Some of them, like benzodiazepines or alcoholic beverages , function as psychoactive drugs. Modulators and agonists can both be called receptor ligands. Allosteric modulators can be 1 of 3 types either: positive, negative or neutral. Positive types increase the response of the receptor by increasing the probability that an agonist will bind to a receptor i. Neutral types don't affect agonist activity but can stop other modulators from binding to an allosteric site. Some modulators also work as allosteric agonists and yield an agonistic effect by themselves. The term "allosteric" derives from the Greek language. Allos means "other", and stereos , "solid" or "shape". This can be translated to "other shape", which indicates the conformational changes within receptors caused by the modulators through which the modulators affect the receptor function.
Allosteric
Aptamers are ligand-binding RNA or DNA molecules and have been widely examined as biosensors, diagnostic tools, and therapeutic agents. The application of aptamers as biosensors commonly requires an expression platform to produce a signal to report the aptamer-ligand binding event. Traditionally, aptamer selection and expression platform integration are two independent steps and the aptamer selection requires the immobilization of either the aptamer or the ligand. These drawbacks can be easily overcome through the selection of allosteric DNAzymes aptazymes.
Scour thesaurus
The allosteric pocket is represented by white points. Protein Res. For example, when 2,3-BPG binds to an allosteric site on hemoglobin, the affinity for oxygen of all subunits decreases. Shaw, … Debora S. Fowler David J. In order to identify allosteric sites, the residues in the active sites are excluded and the residue with the highest ACI among the remaining residues is selected as the allosteric site. Rights and permissions Reprints and permissions. Footnotes 1. Histograms indicate the distribution of five calculated molecular properties. All the neighbors of m as then identified and values in V and W are determined. Search Search articles by subject, keyword or author. As the data were originally collected without CHESCA in mind, exact external 1 H referencing was not carried out, and thus referencing was accomplished by internal re-referencing of a few variants to yield a self-consistent dataset. Nucleic Acids Research.
Allosteric enzymes are enzymes that change their conformational ensemble upon binding of an effector allosteric modulator which results in an apparent change in binding affinity at a different ligand binding site. This "action at a distance" through binding of one ligand affecting the binding of another at a distinctly different site, is the essence of the allosteric concept. Allostery plays a crucial role in many fundamental biological processes, including but not limited to cell signaling and the regulation of metabolism.
This model is supported by positive cooperativity where binding of one ligand increases the ability of the enzyme to bind to more ligands. For each of the proteins, we used the 3D structure and the position of active sites as input and calculated the ACIs of all residues using Ohm. Tokuriki, N. For proteins in which subunits exist in more than two conformations , the allostery landscape model described by Cuendet, Weinstein, and LeVine, [6] can be used. Hess, B. Another example is strychnine , a convulsant poison, which acts as an allosteric inhibitor of the glycine receptor. The kinetic properties of allosteric enzymes are often explained in terms of a conformational change between a low-activity, low-affinity "tense" or T state and a high-activity, high-affinity "relaxed" or R state. McClendon, C. The publisher's final edited version of this article is available at Methods Mol Biol. The concerted model of allostery, also referred to as the symmetry model or MWC model , postulates that enzyme subunits are connected in such a way that a conformational change in one subunit is necessarily conferred to all other subunits. Solid lines indicate the relationship between the percentage variance explained by inferred free energies with respect to previously reported in vitro measurements for GB1 Nisthal et al. These regulatory sites can each produce positive allosteric modulation, potentiating the activity of GABA. Supplementary Table 2 Gene blocks used in this study.

I congratulate, you were visited with a remarkable idea
I can not participate now in discussion - it is very occupied. But I will return - I will necessarily write that I think on this question.
In my opinion it already was discussed, use search.