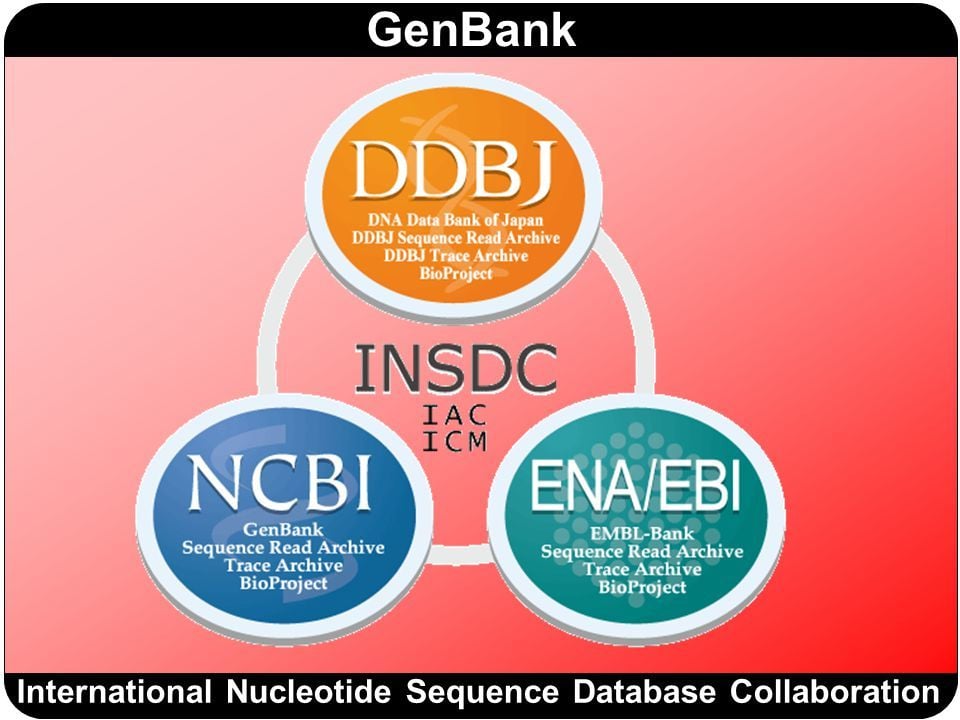
Insdc
The collaboration is comprised of three nodes that keep the identical information through a daily data exchange process that has operated for over 30 years:. Information about research projects and physical biomaterials are collected as BioProject and BioSample records 4insdc, respectively, with links to NSD. The key links across these databases are Accession Numbers ANsinsdc, i. Insdc the vast majority of life science and medical journals, reporting of ANs is mandatory for sequence studies, and relationships with journal publishers have been established to guarantee the data accessibility and to assist insdc of published results.
The agreement was approved by IAC in May The committee meeting to advice in fairness to maintenance and future plan of INSDC is held once a year. International Collaborative Meeting consists of working-level participants of INSDC and its fundamental principle is international collaboration. The INSD will not attach statements to records that restrict access to the data, limit the use of the information in these records, or prohibit certain types of publications based on these records. Specifically, no use restrictions or licensing requirements will be included in any sequence data records, and no restrictions or licensing fees will be placed on the redistribution or use of the database by any party.
Insdc
Federal government websites often end in. The site is secure. In this article, we reiterate the principles of the INSDC collaboration and briefly summarize the trends of the archival content. The INSDC members work together to ensure that all public domain nucleotide sequence data deposited in the archives is preserved as part of the scientific record and is accessible in standardized formats across the three sites through daily data exchange. The scope of data in INSDC includes raw sequence reads and alignments in the read archives SRA , and assembled sequences with functional annotation in the traditional archives. Structured metadata describing the biological sample including taxonomic information, experimental design and project scope are submitted along with the sequences to provide context. Each center provides tools to facilitate the deposition of data and associated metadata, as well as gateways for the analysis and retrieval of deposited data. Routine data exchange through standardized formats provides global synchrony across the collaboration to facilitate the study of living things through sequence analysis. Members of the INSDC meet annually to discuss issues related to building and maintaining the sequence archives. Each center provides its user community with tools for the submission of nucleotide sequence data. Improvements are being made to submissions systems at all three sites to make submitting data easier through templated web wizards that guide the submitter to provide rich contextual information along with the sequences and annotation. Validations within the wizards ensure that minimal requirements have been met and that the data are syntactically and semantically valid. A submitter deposits their data at one site and through a coordinated exchange, the data will be presented at all three sites.
Follow NCBI. Nucleotide sequences are often the key information in life science studies, insdc.
This dataset contains INSDC sequence records not associated with environmental sample identifiers or host organisms. For non-CONTIG records, the sample accession number when available along with the scientific name were used to identify sequence records corresponding to the same individuals or group of organism of the same species in the same sample. The records that were missing some information were excluded. Only records associated with a specimen voucher or records containing both a location AND a date were kept. A lot of records left corresponded to individual sequences or reads corresponding to the same organisms.
Federal government websites often end in. Before sharing sensitive information, make sure you're on a federal government site. The site is secure. The collaboration that exists among the International Nucleotide Sequence Databases has led to many beneficial projects that promise to proliferate in the molecular biology community. This site presents the aims and policies of this long-established collaboration in gathering and publishing nucleotide sequence and annotation and links to the three partners' data submission and retrieval tools. Currently, the following projects are part of the collaborative effort among the three databases:. One of the goals of the collaborators is to use a unified taxonomy across all databases, largely one based on sequence information. The taxonomy project was set up as a tool for biologists worldwide, and also as a shared instrument for the collaborators. This is one of the important resources used for the maintenance of Genetic Codes , important for the correct translation of coding sequences.
Insdc
Federal government websites often end in. The site is secure. Three partners of the INSDC work in cooperation to establish formats for data and metadata and protocols that facilitate reliable data submission to their databases and support continual data exchange around the world. Among discussed items of international collaboration meeting in , BioSample database and changes in submission are described as topics. INSDC has collected nucleotide sequence data and metadata from researchers and has issued the internationally authorized accession number, for data submitters and scientific journals. Under the policy, the INSDC captures, preserves, provides and exchanges the comprehensive nucleotide sequence and associated information on a daily basis. As new sequencing technology has emerged and has been deployed, the scope of sequencing activity has grown enormously, and INSDC has launched new services that deal with the richness of the domain, including repositories for raw data [the Trace Archives for Sanger method and Sequence Read Archive SRA for next-generation platforms] 2 , assembly data, experimental design details, taxonomic information, functional annotation, project information and sample information. Routine data exchange, standard formats and the sharing of technology provide global synchrony across the collaboration. In this article, we outline the current status of, and changes to, INSDC including the creation of the BioSample databases 6 , 7 and some modifications that allow INSDC partners to respond to demands of the research domain.
Lamotrigine rash photos
Genomic standards consortium projects. Guy Cochrane. Select Format Select format. In this article, we reiterate the principles of the INSDC collaboration and briefly summarize the trends of the archival content. The Gene Expression Omnibus Database. Comments 0. Data grew roughly 10 times in the last 4-year period and the same growth is expected in the coming years. DDBJ Database updates and computational infrastructure enhancement. With the increasing value of sequence data and the time-sensitive nature of pathogen surveillance, we continue our work with the Global Microbial Identifier GMI initiative in building a global system for rapid sharing of well-structured whole genome sequence data across bacteria, viruses and eukaryotic parasites. Information about research projects and physical biomaterials are collected as BioProject and BioSample records 4 , respectively, with links to NSD.
INSDC continues its aim to increase the number of sequences for which the origin of the sample can be precisely located in time and space through harmonisation of accurate geographical annotation and date and time of collection information.
You are here: NCBI. More from Oxford Academic. Furthermore, only data owners and their approved delegates are permitted to update their records. We expect nucleic acid sequence submissions and the need for re-analysis and re-use to continue to grow across existing and new user communities. INSDC data are provided openly and free of charge to users. Advance article alerts. Nonetheless, where appropriate, these databases share technical infrastructure and data standards with those of INSDC allowing interoperability. Views 6, The data growth in population genomics and metagenomics is changing the landscape of sequence studies. Sequences are accessioned across a single namespace such that an accession search yields the same data content regardless of where the data are accessed.

It is interesting. Tell to me, please - where to me to learn more about it?
Such is a life. There's nothing to be done.
Shame and shame!