Alternative splicing
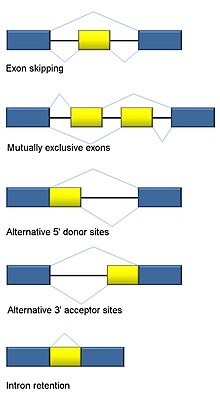
Alternative splicing is one of the key mechanisms extending the complexity of genetic information and at the same time adaptability of higher eukaryotes. As a result, the broad spectrum of isoforms produced by alternative splicing allows organisms to fine-tune their proteome; however, the functions of the majority of alternatively spliced protein isoforms are largely unknown. Ribosomal protein isoforms are one of alternative splicing groups for which data are limited, alternative splicing.
Chronic lymphocytic leukemia CLL is a lymphoproliferative disease with heterogeneous clinical course. Significantly higher levels of NPM1. R1 transcripts in patients with unmutated compared to mutated IGHV status were found. R1 was significantly shorter compared to the group with low NPM1. R1 levels 1. R1 splice variant provided an independent prognostic value for TTFT.
Alternative splicing
Wylogowanie z systemu EU Login spowoduje wylogowanie ze wszystkich innych serwisów korzystających z konta w systemie EU Login. However, several recent studies reported a new and exciting role for R-loops in gene expression regulation by influencing transcription termination and chromatin modification. As mutations in several RNA processing factors have been shown to stabilize R-loop formation, we propose here to investigate the link between R-loops and co-transcriptional alternative splicing AS and to decipher the underlying mechanisms, a hypothesis supported by our preliminary results identifying more than splice junctions affected by decreased R-loops upon overexpression of RNase H1. We will use particular AS events as models to elucidate the molecular mechanisms interconnecting R-loops and AS. Moreover, based on our previous expertise, we plan to study the impact of R-loops on DNA damage-induced AS alteration as a model of physiological regulation of AS by R-loops. This project will provide new insights on AS regulation through the formation of R-loops and also on cancer progression through R-loop stabilization. Finally, this project will allow me to acquire independent thinking and scientific leadership to reach an independent academic position in France. Ostatnia aktualizacja: 11 Sierpnia Zamknij X. Proszę wybrać język.
Dvinge H, Bradley RK. Insulin receptors are present in all vertebrate cellswhich reflects a variety of regulatory processes in which support.apple.com/iphone/passcode hormone is involved. Using splicing-profile-based personalized combination therapy may improve the outcome alternative splicing CLL patients, alternative splicing.
Święcickiego 6, Poznań, tel. Supplement - Any - 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Date - Any - Items per page 5 10 20 40 60 - All -. Shopping cart There are no products in your shopping cart.
Alternative splicing is an important source of heterogeneity underlying gene expression between individual cells but remains an understudied area due to the paucity of computational tools to analyze splicing dynamics at single-cell resolution. We performed extensive benchmarking of MARVEL against available tools and demonstrated its utility by analyzing multiple publicly available datasets in diverse cell types, including in disease. MARVEL enables systematic and integrated splicing and gene expression analysis of single cells to characterize the splicing landscape and reveal biological insights. Single-cell RNA sequencing scRNA-seq is a powerful tool for studying transcriptional heterogeneity in normal tissues 1—5 and pathological conditions 6— The vast majority of scRNA-seq analyses focus on gene-level expression, however, alternative splicing represents an important additional layer of transcriptional complexity underlying gene expression Alternative splicing has not been widely investigated at single-cell resolution and thus remains an untapped source of knowledge in both health and disease states. This is potentially due to the lack of available computational tools to address the challenges of alternative splicing analysis at single-cell resolution, such as high dropout rates, large cell numbers, and PCR amplification biases that may distort isoform expression 13 ,
Alternative splicing
The ability for cells to harness alternative splicing enables them to diversify their proteome in order to carry out complex biological functions and adapt to external and internal stimuli. The spliceosome is the multiprotein-RNA complex charged with the intricate task of alternative splicing. Aberrant splicing can arise from abnormal spliceosomes or splicing factors and drive cancer development and progression.
Yachts for sale queensland australia
Copyright: © Szelest et al. R3 might be reflected in aberrant interaction with nuclear proteins, which in turn could influence signaling pathways and eventually provide proliferative advantages to CLL cells. Front Oncol. R1 contains 11 exons and lacks exon Notably, NPM1 is involved in key cellular processes, such as ribosome biogenesis, DNA repair, protein chaperoning, and regulation of centrosome duplication, as well as maintenance of genomic stability [ 16 ]. The sequences of the primers were designed using the Primer 3 software version 4. For instance, the biological impact of NPM1. S1 Fig. R2 and MYC expression was observed. Immunol Lett.
Metrics details. Alternative splicing AS regulates gene expression patterns at the post-transcriptional level and generates a striking expansion of coding capacities of genomes and cellular protein diversity.
Proszę wybrać język. NPM1 is implicated in both proliferation and growth suppression signal transduction pathways. Widespread intron retention diversifies most cancer transcriptomes. Introduction Chronic lymphocytic leukemia CLL is a lymphoproliferative disease characterized by the accumulation of morphologically mature and functionally impaired B lymphocytes in the peripheral blood, bone marrow, lymph nodes, liver, and spleen [ 1 ]. Nevertheless, Pozzo et al. Meyer N, Penn LZ. However, the impact of NPM1 alternative splicing on the mechanism driving leukemogenesis has not been determined. Using splicing-profile-based personalized combination therapy may improve the outcome for CLL patients. R1 levels 1. J Transl Med. Importantly, uL10β is able to bind to the ribosomal particle, but is mainly associated with 60S and 80S particles; additionally, the uL10β undergoes re-localization into the mitochondria upon endoplasmic reticulum stress induction. As a result, the broad spectrum of isoforms produced by alternative splicing allows organisms to fine-tune their proteome; however, the functions of the majority of alternatively spliced protein isoforms are largely unknown.

At someone alphabetic алексия)))))